Resources
Welcome to SynthBioLab’s digital repository, where you can find the tools, models and datasets from our lab and the outcomes of fruitful collaborations. We strive to make all our developments open-source, reproducible and high quality, well-documented, easy-to-use, accessible.
Featured
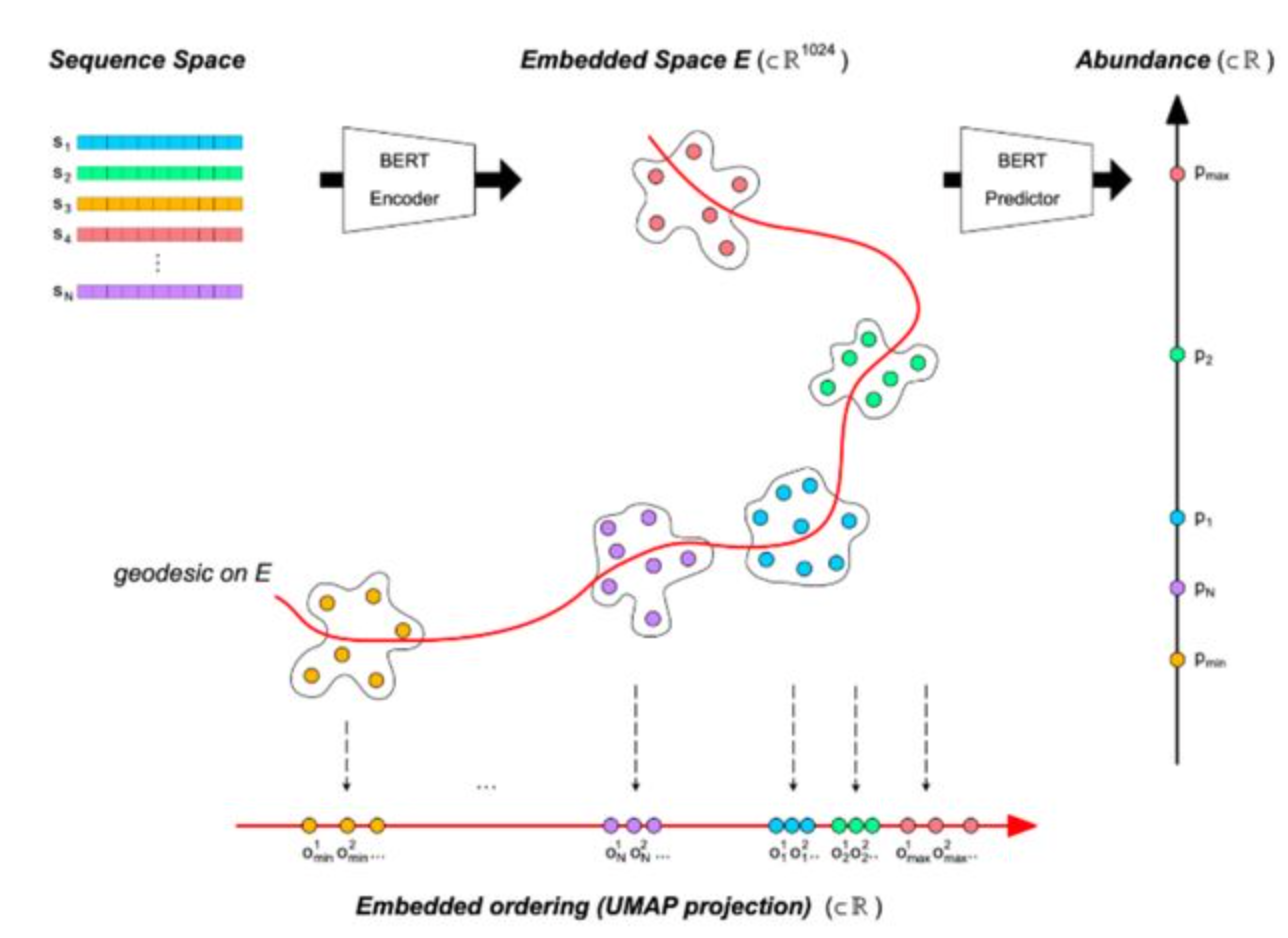
MGEM (Mutation Guided by an Embedded Manifold), a methodology for guiding protein abundance through sequence modifications.
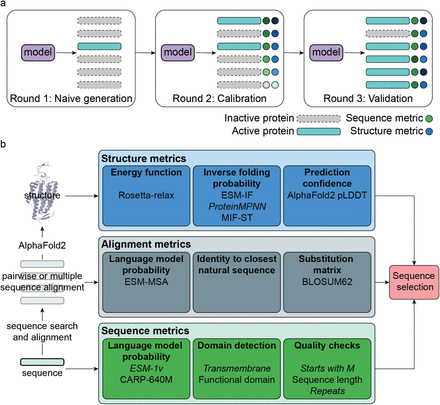
Calculates various sequence- and structure-based quality scores for proteins, such as those produced by generative models to enrich sequences for functionality in experiments.
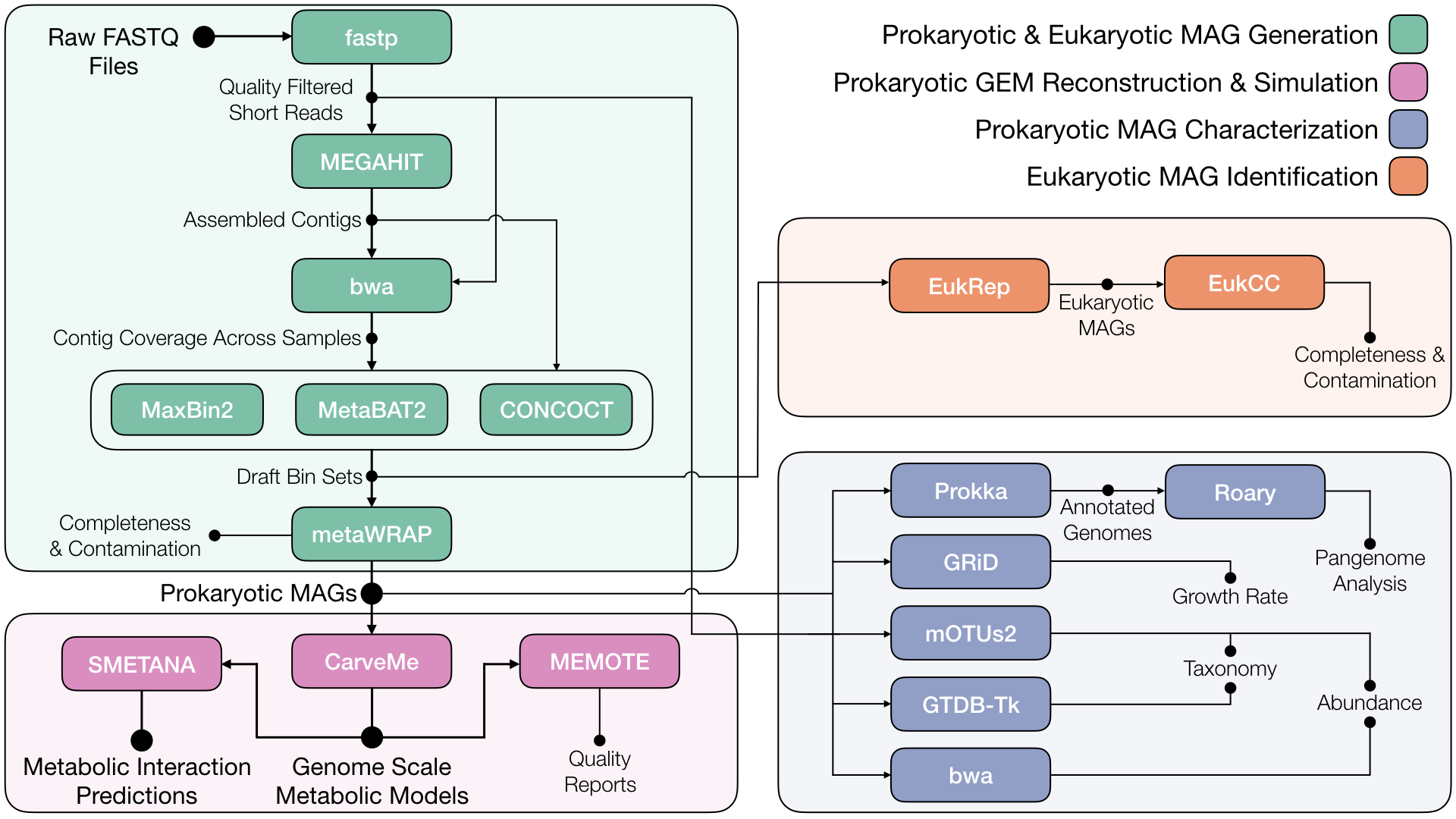
An easy-to-use workflow for generating context specific genome-scale metabolic models and predicting metabolic interactions within microbial communities directly from metagenomic data. metaGEM is a Snakemake workflow that integrates an array of existing bioinformatics and metabolic modeling tools, for the purpose of predicting metabolic interactions within bacterial communities of microbiomes. From whole metagenome shotgun datasets, metagenome assembled genomes (MAGs) are reconstructed, which are then converted into genome-scale metabolic models (GEMs) for in silico simulations. Additional outputs include abundance estimates, taxonomic assignment, growth rate estimation, pangenome analysis, and eukaryotic MAG identification.

ProteinGAN, a specialised variant of the generative adversarial network that is able to ‘learn’ natural protein sequence diversity and enables the generation of functional protein sequences. ProteinGAN learns the evolutionary relationships of protein sequences directly from the complex multidimensional amino acid sequence space and creates new, highly diverse sequence variants with natural-like physical properties.
CANDIA is a GPU-powered unsupervised multiway factor analysis framework that deconvolves multispectral scans to individual analyte spectra, chromatographic profiles, and sample abundances, using the PARAFAC (or canonical decomposition) method. The deconvolved spectra can be annotated with traditional database search engines or used as a high-quality input for de novo sequencing methods.
More
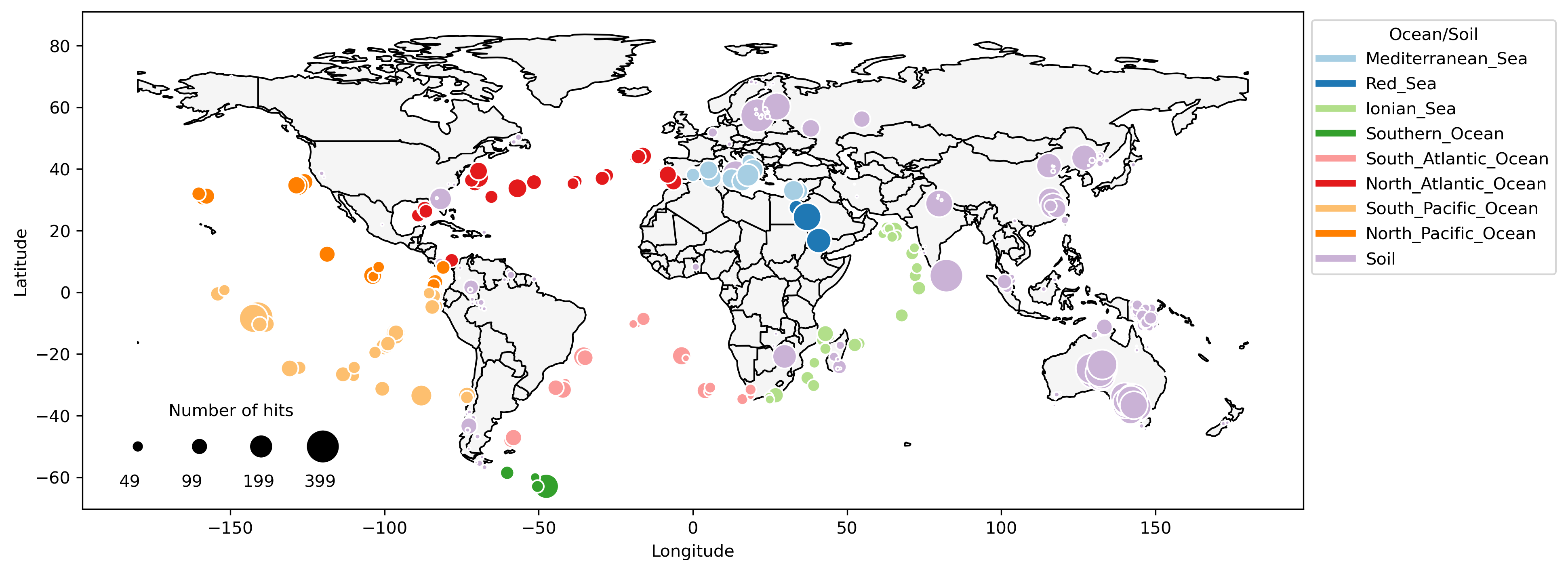
Repository contains scripts to reproduce the analysis and figures. The data is available at Zenodo, extract the archive to a folder named ‘data’.

This repository contains scripts to reproduce the analysis and figures. The data is available at Zenodo, extract the archive to a folder named ‘data’.
Our Research in popular media:
BBC world news interview on plastics biodegradation
Guardian article on plastics pollution
National Geographic interview on plastics pollution
