Research
Full up to date-list of publications is available on Google Scholar.
Highlighted
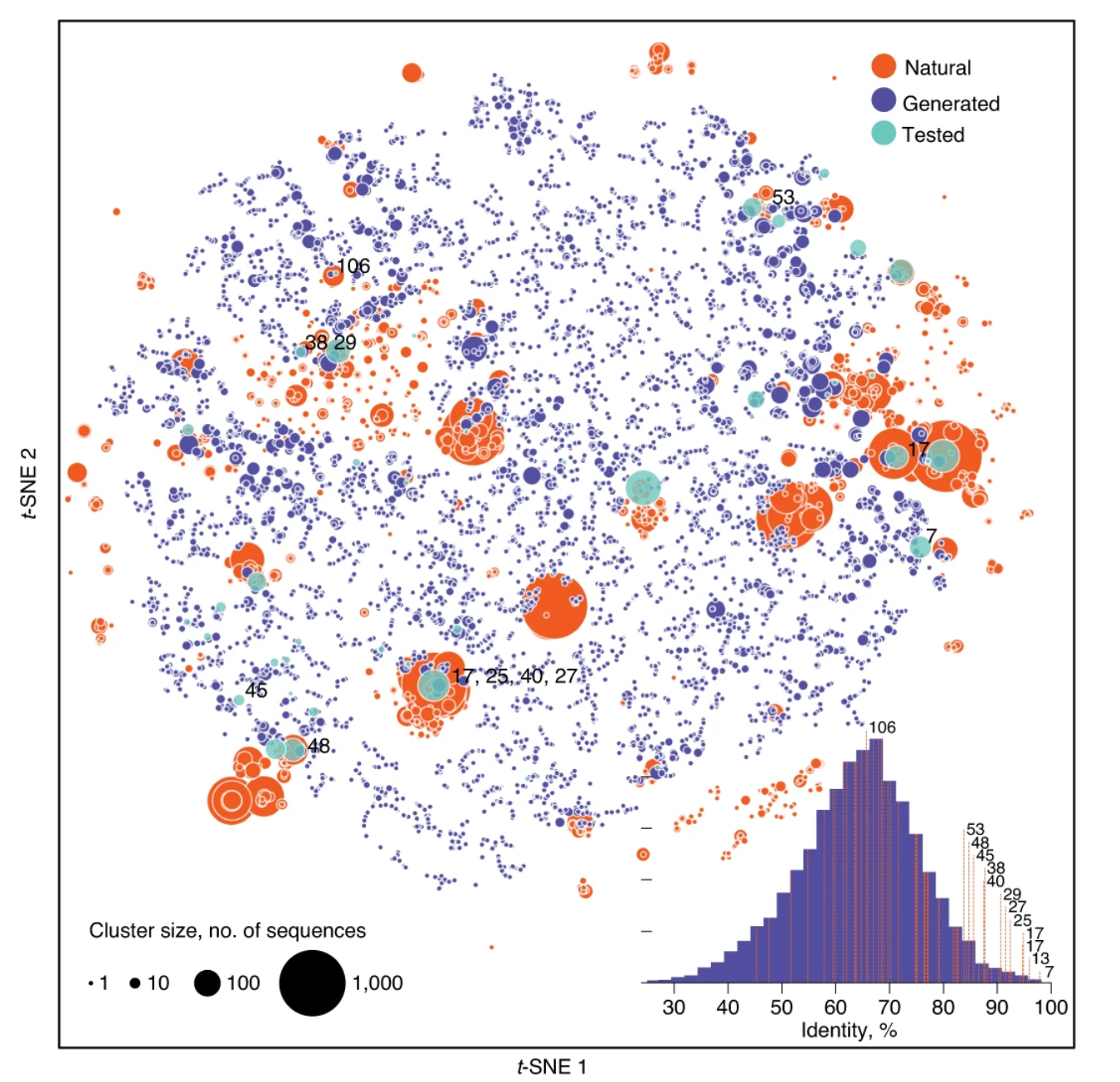
Expanding functional protein sequence spaces using generative adversarial networks
Nature Machine Intelligence
·
04 Mar 2021
·
doi:10.1038/s42256-021-00310-5
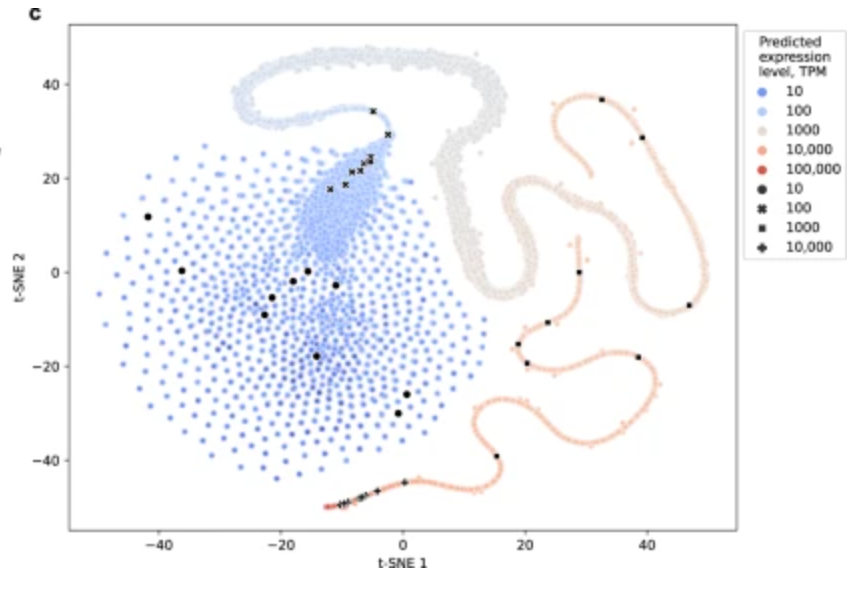
Controlling gene expression with deep generative design of regulatory DNA
Nature Communications
·
30 Aug 2022
·
doi:10.1038/s41467-022-32818-8

Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure
Nature Communications
·
01 Dec 2020
·
doi:10.1038/s41467-020-19921-4
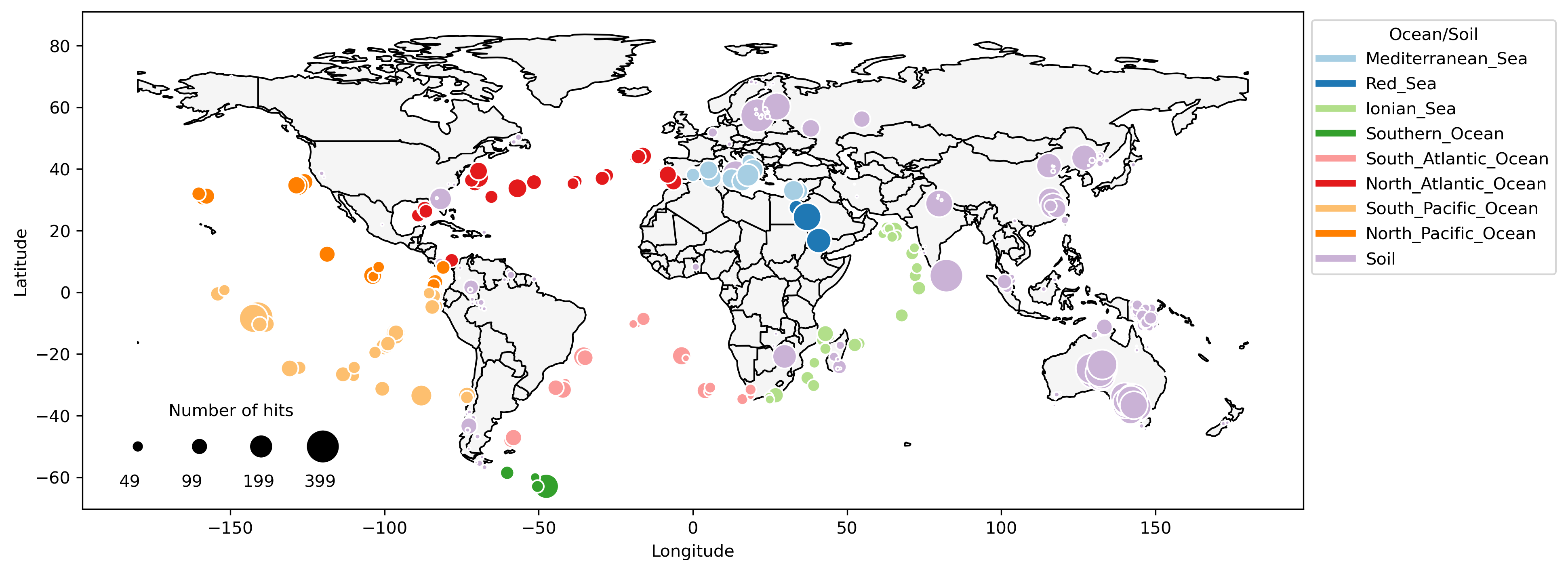
Plastic-Degrading Potential across the Global Microbiome Correlates with Recent Pollution Trends
mBio
·
26 Oct 2021
·
doi:10.1128/mbio.02155-21
All
2024
Single Distal Mutation Enhances Activity of known PETases via Stabilisation of PET-binding
Cold Spring Harbor Laboratory
·
11 Sep 2024
·
doi:10.1101/2024.09.11.612432
Author Correction: Computational scoring and experimental evaluation of enzymes generated by neural networks
Nature Biotechnology
·
20 May 2024
·
doi:10.1038/s41587-024-02275-3
Computational scoring and experimental evaluation of enzymes generated by neural networks
Nature Biotechnology
·
23 Apr 2024
·
doi:10.1038/s41587-024-02214-2
2023
The amino acid sequence determines protein abundance through its conformational stability and reduced synthesis cost
Cold Spring Harbor Laboratory
·
04 Oct 2023
·
doi:10.1101/2023.10.02.560091
Synthesis of Metal–Organic Frameworks through Enzymatically Recycled Polyethylene Terephthalate
ACS Sustainable Chemistry & Engineering
·
21 Sep 2023
·
doi:10.1021/acssuschemeng.3c05222
Computational Scoring and Experimental Evaluation of Enzymes Generated by Neural Networks
Cold Spring Harbor Laboratory
·
04 Mar 2023
·
doi:10.1101/2023.03.04.531015
The Impact of Acute Nutritional Interventions on the Plasma Proteome
The Journal of Clinical Endocrinology & Metabolism
·
20 Jan 2023
·
doi:10.1210/clinem/dgad031
Cell-cell metabolite exchange creates a pro-survival metabolic environment that extends lifespan
Cell
·
01 Jan 2023
·
doi:10.1016/j.cell.2022.12.007
2022
Toward learning the principles of plant gene regulation
Trends in Plant Science
·
01 Dec 2022
·
doi:10.1016/j.tplants.2022.08.010
Learning deep representations of enzyme thermal adaptation
Protein Science
·
22 Nov 2022
·
doi:10.1002/pro.4480

Controlling gene expression with deep generative design of regulatory DNA
Nature Communications
·
30 Aug 2022
·
doi:10.1038/s41467-022-32818-8
Data mining of Saccharomyces cerevisiae mutants engineered for increased tolerance towards inhibitors in lignocellulosic hydrolysates
Biotechnology Advances
·
01 Jul 2022
·
doi:10.1016/j.biotechadv.2022.107947
Enhanced metabolism and negative regulation of ER stress support higher erythropoietin production in HEK293 cells
Cell Reports
·
01 Jun 2022
·
doi:10.1016/j.celrep.2022.110936
A proteomic survival predictor for COVID-19 patients in intensive care
PLOS Digital Health
·
18 Jan 2022
·
doi:10.1371/journal.pdig.0000007
2021

Plastic-Degrading Potential across the Global Microbiome Correlates with Recent Pollution Trends
mBio
·
26 Oct 2021
·
doi:10.1128/mbio.02155-21
metaGEM: reconstruction of genome scale metabolic models directly from metagenomes
Nucleic Acids Research
·
06 Oct 2021
·
doi:10.1093/nar/gkab815
A time-resolved proteomic and prognostic map of COVID-19
Cell Systems
·
01 Aug 2021
·
doi:10.1016/j.cels.2021.05.005
Potential for improved retention rate by personalized antiseizure medication selection: A register‐based analysis
Epilepsia
·
09 Jul 2021
·
doi:10.1111/epi.16987
Learning the Regulatory Code of Gene Expression
Frontiers in Molecular Biosciences
·
10 Jun 2021
·
doi:10.3389/fmolb.2021.673363
Ultra-fast proteomics with Scanning SWATH
Nature Biotechnology
·
25 Mar 2021
·
doi:10.1038/s41587-021-00860-4

Expanding functional protein sequence spaces using generative adversarial networks
Nature Machine Intelligence
·
04 Mar 2021
·
doi:10.1038/s42256-021-00310-5
Benchmarking accuracy and precision of intensity‐based absolute quantification of protein abundances in Saccharomyces cerevisiae
PROTEOMICS
·
23 Feb 2021
·
doi:10.1002/pmic.202000093
Bayesian genome scale modelling identifies thermal determinants of yeast metabolism
Nature Communications
·
08 Jan 2021
·
doi:10.1038/s41467-020-20338-2
2020
Parallel Factor Analysis Enables Quantification and Identification of Highly Convolved Data-Independent-Acquired Protein Spectra
Patterns
·
01 Dec 2020
·
doi:10.1016/j.patter.2020.100137

Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure
Nature Communications
·
01 Dec 2020
·
doi:10.1038/s41467-020-19921-4
Transcriptome analysis of EPO and GFP HEK293 Cell-lines Reveal Shifts in Energy and ER Capacity Support Improved Erythropoietin Production in HEK293F Cells
Cold Spring Harbor Laboratory
·
16 Sep 2020
·
doi:10.1101/2020.09.16.299966
Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection
Cell Systems
·
01 Jul 2020
·
doi:10.1016/j.cels.2020.05.012
Parallel factor analysis enables quantification and identification of highly-convolved data independent-acquired protein spectra
Cold Spring Harbor Laboratory
·
23 Apr 2020
·
doi:10.1101/2020.04.21.052654
Benchmarking accuracy and precision of intensity-based absolute quantification of protein abundances inSaccharomyces cerevisiae
Cold Spring Harbor Laboratory
·
24 Mar 2020
·
doi:10.1101/2020.03.23.998237
2018
Machine Learning Predicts the Yeast Metabolome from the Quantitative Proteome of Kinase Knockouts
Cell Systems
·
01 Sep 2018
·
doi:10.1016/j.cels.2018.08.001
Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies
Nature Microbiology
·
19 Mar 2018
·
doi:10.1038/s41564-018-0123-9
Cost-effective generation of precise label-free quantitative proteomes in high-throughput by microLC and data-independent acquisition
Scientific Reports
·
12 Mar 2018
·
doi:10.1038/s41598-018-22610-4
The Response to Past Climate Perturbations Explains Extremely Low Genetic Diversity in the Genome of an Abundant Ice-Age Remnant, the Alpine Marmot
SSRN Electronic Journal
·
01 Jan 2018
·
doi:10.2139/ssrn.3219259
2017
The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization
Nature Communications
·
10 Jul 2017
·
doi:10.1038/ncomms16018
2016
Functional Metabolomics Describes the Yeast Biosynthetic Regulome
Cell
·
01 Oct 2016
·
doi:10.1016/j.cell.2016.09.007
Precise label-free quantitative proteomes in high-throughput by microLC and data-independent SWATH acquisition
Cold Spring Harbor Laboratory
·
05 Sep 2016
·
doi:10.1101/073478
The metabolic background is a global player in Saccharomyces gene expression epistasis
Nature Microbiology
·
01 Feb 2016
·
doi:10.1038/nmicrobiol.2015.30
2015
Metabolic dependencies drive species co-occurrence in diverse microbial communities
Proceedings of the National Academy of Sciences
·
04 May 2015
·
doi:10.1073/pnas.1421834112
2014
Contribution of Network Connectivity in Determining the Relationship between Gene Expression and Metabolite Concentration Changes
PLoS Computational Biology
·
24 Apr 2014
·
doi:10.1371/journal.pcbi.1003572
2012
Prediction and identification of sequences coding for orphan enzymes using genomic and metagenomic neighbours
Molecular Systems Biology
·
01 Jan 2012
·
doi:10.1038/msb.2012.13
2011
Flux coupling and transcriptional regulation within the metabolic network of the photosynthetic bacterium Synechocystis sp. PCC6803
Biotechnology Journal
·
11 Jan 2011
·
doi:10.1002/biot.201000109
2010
Metabolic Network Topology Reveals Transcriptional Regulatory Signatures of Type 2 Diabetes
PLoS Computational Biology
·
01 Apr 2010
·
doi:10.1371/journal.pcbi.1000729
